Science Strategies
TACF Strategic Science Plan
2023-2033
During the past 100 years, chestnut blight, caused by the Asian fungus Cryphonectria parasitica, killed an estimated four billion American chestnut (Castanea dentata) trees, and pushed the iconic species into a state of functional extinction. Human interference triggered the American chestnut’s demise–now scientific innovation offers us the best chance to save it. As reliable and productive as the American chestnut tree was, the species now exists in forests primarily in a state of stunted vegetative regeneration from surviving root systems, and this population continues to dwindle. The American Chestnut Foundation (TACF) is leading an unprecedented rescue mission to return the iconic American chestnut to its native range.
Our Major Goals
Resistance
To develop American chestnut trees with sufficient resistance to two deadly diseases. As a first priority, we focus on chestnut blight. Our second priority will be resistance to Phytophthora root rot (PRR) caused by the non-native and invasive soil pathogen Phytophthora cinnamomi.
Diversity
To characterize, preserve, and utilize the genetic diversity of existing American chestnut trees for research and to ensure that the developed disease resistance is incorporated into environmentally-adaptable, diverse populations.
Restoration
To promote forest plantings of genetically diverse and disease-resistant American chestnuts capable of sustained population growth and expansion across the broad and ever-changing landscape of our Eastern hardwood forests.
Current State of TACF’s Research and Breeding Program
To achieve its mission, TACF employs a multi-pronged and adaptive program of research and development informed by staff and volunteer scientists as well as research collaborators in universities and other organizations. TACF’s Director of Science has brought the power of genomics—the study of the genetic material of an organism via molecular approaches—to support all our future efforts including breeding. Collaboration with the Hudson-Alpha Institute of Biology has resulted in the complete DNA sequencing of the genomes of one American and two Chinese chestnut trees. In the six years since our 2017 Strategic Plan, we have made extensive use of this technology and also improved methods for evaluating and rating resistance to both chestnut blight and PRR.
Breeding for resistance to disease requires methods of accurately assessing the resistance of individual trees. In the past several years, TACF has developed and tested improved methods for evaluating and rating trees for blight resistance. These include the use of small stem assays (SSAs) performed on potted seedlings, improved phenotype scoring methods for field-grown trees, and the use of genomic prediction models for scoring resistance based on genotype. Together, these approaches now allow us to screen more trees, more rapidly and more accurately than in the past. TACF and its partners have developed methods for screening seedlings for PRR resistance and we have now established a screening facility for PRR resistance in cooperation with the U.S. Forest Service at their Bent Creek southern research station in Asheville, NC.
Thirty years of backcross breeding to capture the natural blight resistance of Chinese chestnut, involving over 50,000 trees, has resulted in approximately 500 trees that contain on average only 12% Chinese chestnut DNA but have inherited at least some blight resistance, and some of these advanced hybrids also exhibit the desired character typical of American chestnuts. However, the overall number and the degree of resistance of trees coming out of the backcross program is insufficient for use as a restoration population without further breeding efforts. Recent genetic analysis confirms that blight resistance is a complex process controlled by many more than two or three genes as previously hypothesized. This explains the modest degree of success, and points to the need for a different breeding strategy. Two have been identified as having near-term potential.
One promising approach is to build upon the backcross program with recurrent selection, beginning with the most resistant backcross hybrids. Computer modelling suggests that this approach can substantially increase the mean level of blight resistance in our trees, and we are already testing this prediction through the creation of “best x best” crosses. We anticipate that it will take 2-3 years to generate enough data to demonstrate whether the best x best approach is likely to produce desired results.
TACF’s second near-term approach involves the use of transgenic technology. The most promising method involves inserting a wheat gene that encodes an enzyme, oxalate oxidase (OxO), that degrades oxalic acid, the toxin by which the pathogen kills. The Darling line, developed at SUNY-ESF in 2016, used the 35S constitutive promoter to allow constant expresssion of the gene, but years of testing revealed varying levels of blight resistance, higher mortality, and a suite of performance issues likely linked to metabolic costs from high constitutive expression of the transgene. TACF and other research partners are now exploring transgenic lines that use a variety of inducible promoters to express the OxO gene only under specific conditions, such as in the tissue surrounding a wound where the blight fungus is commonly introduced.
TACF also considers it essential to incorporate resistance to PRR into all its breeding efforts in order to have resistant material adapted to all regions where P. cinnamomi may spread. Although PRR is of regional importance in the southern part of the chestnut’s range, it is a growing problem in previously unaffected central and northern areas. Our population of backcross hybrids includes trees with high levels of American ancestry but which also carry PRR-resistance from their Chinese ancestors. Unlike for chestnut blight, genomic analysis has identified a small number of specific regions in the Chinese chestnut genome that are strongly associated with resistance to PRR. This likely explains our faster progress in capturing PRR resistance, and also gives us confidence that genomic selection will advance progress in creating PRR-resistant populations.
To achieve its mission, TACF employs a multi-pronged and adaptive program of research and development informed by staff and volunteer scientists as well as research collaborators in universities and other organizations. TACF’s Director of Science has brought the power of genomics—the study of the genetic material of an organism via molecular approaches—to support all our future efforts including breeding. Collaboration with the Hudson-Alpha Institute of Biology has resulted in the complete DNA sequencing of the genomes of one American and two Chinese chestnut trees. In the six years since our 2017 Strategic Plan, we have made extensive use of this technology and also improved methods for evaluating and rating resistance to both chestnut blight and PRR.
Breeding for resistance to disease requires methods of accurately assessing the resistance of individual trees. In the past several years, TACF has developed and tested improved methods for evaluating and rating trees for blight resistance. These include the use of small stem assays (SSAs) performed on potted seedlings, improved phenotype scoring methods for field-grown trees, and the use of genomic prediction models for scoring resistance based on genotype. Together, these approaches now allow us to screen more trees, more rapidly and more accurately than in the past. TACF and its partners have developed methods for screening seedlings for PRR resistance and we have now established a screening facility for PRR resistance in cooperation with the U.S. Forest Service at their Bent Creek southern research station in Asheville, NC.
Thirty years of backcross breeding to capture the natural blight resistance of Chinese chestnut, involving over 50,000 trees, has resulted in approximately 500 trees that contain on average only 12% Chinese chestnut DNA but have inherited at least some blight resistance, and some of these advanced hybrids also exhibit the desired character typical of American chestnuts. However, the overall number and the degree of resistance of trees coming out of the backcross program is insufficient for use as a restoration population without further breeding efforts. Recent genetic analysis confirms that blight resistance is a complex process controlled by many more than two or three genes as previously hypothesized. This explains the modest degree of success, and points to the need for a different breeding strategy. Two have been identified as having near-term potential.

One promising approach is to build upon the backcross program with recurrent selection, beginning with the most resistant backcross hybrids. Computer modelling suggests that this approach can substantially increase the mean level of blight resistance in our trees, and we are already testing this prediction through the creation of “best x best” crosses. We anticipate that it will take 2-3 years to generate enough data to demonstrate whether the best x best approach is likely to produce desired results.
TACF’s second near-term approach involves the use of transgenic technology. The most promising method involves inserting a wheat gene that encodes an enzyme, oxalate oxidase (OxO), that degrades oxalic acid, the toxin by which the pathogen kills. The Darling line, developed at SUNY-ESF in 2016, used the 35S constitutive promoter to allow constant expresssion of the gene, but years of testing revealed varying levels of blight resistance, higher mortality, and a suite of performance issues likely linked to metabolic costs from high constitutive expression of the transgene. TACF and other research partners are now exploring transgenic lines that use a variety of inducible promoters to express the OxO gene only under specific conditions, such as in the tissue surrounding a wound where the blight fungus is commonly introduced.
TACF also considers it essential to incorporate resistance to PRR into all its breeding efforts in order to have resistant material adapted to all regions where P. cinnamomi may spread. Although PRR is of regional importance in the southern part of the chestnut’s range, it is a growing problem in previously unaffected central and northern areas. Our population of backcross hybrids includes trees with high levels of American ancestry but which also carry PRR-resistance from their Chinese ancestors. Unlike for chestnut blight, genomic analysis has identified a small number of specific regions in the Chinese chestnut genome that are strongly associated with resistance to PRR. This likely explains our faster progress in capturing PRR resistance, and also gives us confidence that genomic selection will advance progress in creating PRR-resistant populations.
Germplasm Characterization and Preservation
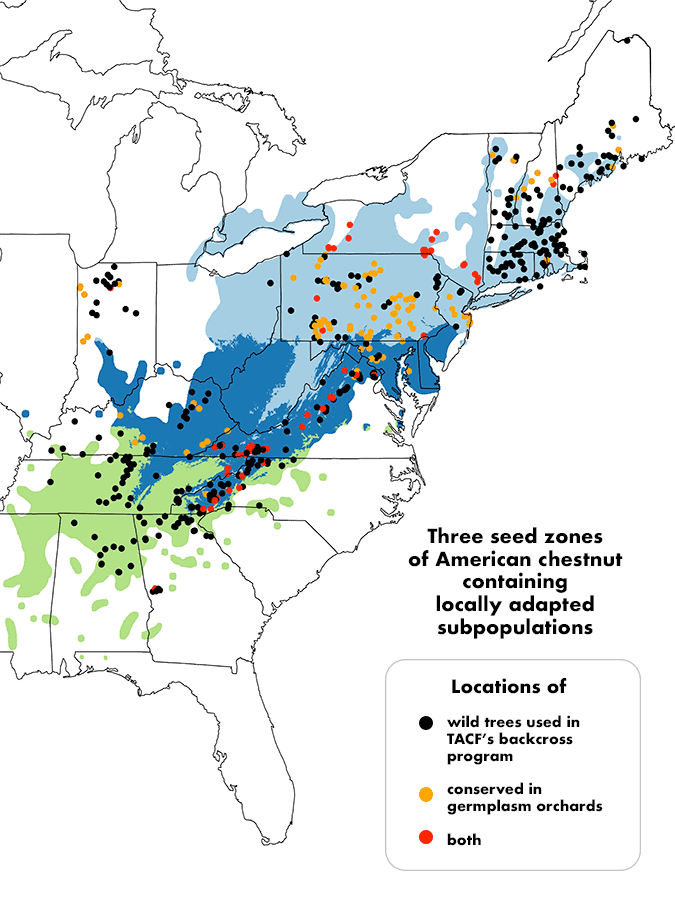
TACF’s collaboration with Virginia Tech on “landscape genomics” research has produced DNA sequence information for hundreds of wild American chestnut trees from Maine to Mississippi. Results show that the existing American chestnut population can be subdivided into three geographic sub-populations—northeastern, central, and southwestern. In addition to providing valuable leads toward future work on genes that control adaptation to climate, the southwestern sub-population has been highlighted for preservation as it is both the most diverse, and the most threatened with extinction.
Preservation of germplasm for research and breeding efforts is challenging. Our volunteers and staff have made impressive efforts to conserve hundreds of wild American chestnut trees in germplasm conservation orchards, but we find that these eventually become threatened by blight as the trees mature. We will continue to replant our most valuable germplasm in new orchards while we explore other conservation approaches, such as freezing pollen, grafting, and when permissible, crossing wild type Americans with transgenic trees to preserve germplasm.
TACF’s backcross population also serves as a reservoir of American chestnut germplasm, conserved in readily accessible orchards throughout the chestnut’s native range. Thus, even backcross trees with relatively low levels of resistance to chestnut blight are important sources of genetic diversity for both transgenic and conventional breeding efforts.
Major Strategic Research Goals
Assess current hybrids and the prospects for continued breeding efforts
TACF is in its second year of testing best x best crosses to confirm whether this approach delivers the expected increase in mean blight resistance. We anticipate that it will take 2-3 more years to obtain data that will clearly inform us how to proceed with conventional breeding.
Determine whether inducible-OxO transgenic American chestnut lines are suitable for development of a restoration population
Insertion of the OxO gene has shown promise for blight resistance, but study of lower expressors of OxO and/or use of other promoters is warranted. If results are promising, TACF will continue to establish seed orchards of blight-resistant transgenic American chestnuts crossed with diverse, regionally adapted American chestnuts. TACF will also test the stacking of resistance through crosses between OxO lines and moderately blight-resistant backcross trees to combine the blight resistance of transgenic trees with the PRR resistance found in some of TACF’s backcross breeding lines. Clearly, such efforts are necessary to determine if OxO transgenic lines will be sufficiently robust for restoration efforts.
Ensure that disease resistance is integrated into environmentally adaptable, genetically diverse germplasm
Regardless of which approaches are used to incorporate disease resistance in our restoration populations, it will eventually be necessary to ensure that there is enough genetic diversity in our populations to avoid inbreeding depression and ensure that populations can continue adapting to variations in growing conditions throughout the chestnut’s native range, as well as to climate change. This will involve continued genomic analysis of our breeding populations, as well as deliberate efforts to conserve American chestnut germplasm, whether through grafting, cryostorage of pollen or crossing with blight-resistant transgenic trees.
Support research into naturally occurring and novel forms of resistance
The strategies above all build on prior accomplishments by TACF and its partners. Even as we pursue them, we recognize the potential for new methods or technologies that accelerate and advance our work. The identification of specific Chinese or American chestnut genes, genomic markers, or metabolites associated with blight resistance could greatly accelerate our breeding program or improve the quality of our trees. The “reference” genomes TACF and its partners have developed are critically important new tools for gene discovery.
This type of work is knowledge-, resource-, and labor-intensive, and we are broadening our partnerships to expand projects aimed at better understanding the molecular relationships between the host and its chestnut blight and PRR pathogens, with a view to developing novel forms of resistance. Currently, TACF has expanded its partnership with scientists at major universities, research organizations, and private companies to identify promising candidate genes for resistance. One current approach correlates the results of genomic mapping of resistance loci with patterns of gene expression of both pathogens and Chinese vs American chestnut hosts during the course of infection. Other approaches involve chemical analyses of changes in metabolites and structural changes related to host defense. TACF will continue to provide seed money to test new approaches, partly through its external grants program as well as seeding new partnerships.
Address major barriers to research on chestnut
Like many hardwood trees, chestnuts pose challenges that, if solved, could speed up progress considerably. None of these challenges has a simple solution, but we nevertheless propose to continually look for new approaches that can help overcome these barriers.
One of these is the long generation time—approximately 7 years from germination to the production of the first male or female flowers. The trees are self-incompatible, meaning they require pollen from another tree for to produce viable seed. This is a valuable trait in nature but not for a breeder in a hurry for results. A major breakthrough is that we can now produce pollen from 1- to 2-year-old seedlings if they are grown under high light, but we still must wait 7 years for female flowers to produce progeny. Manipulation of genes controlling juvenility and flowering might be one of several approaches worth future exploration.
Second, chestnuts, like humans, are notably heterozygous for most genes, meaning that there is great variation between the DNA of the two parents. This means that the progeny will not be identical to their parents but rather represent new mixtures of maternal and paternal genes. If we want to preserve and propagate a very special tree in our orchards, or to experimentally test it for blight resistance in a replicated trial, we need to find a way to clonally propagate valuable trees. Identical genetic clones are usually produced by inducing cuttings to form roots. But to date, chestnuts cannot be persuaded to predictably form roots. We have initiated a new partnership to test the use of Agrobacterium rhizogenes to induce root formation. We have invested staff time in improving our ability to create clones through grafting, but it remains labor-intensive, and the preserved trees have foreign roots. Clonal material can also be propagated through propagation of embryos in tissue culture and then inducing them to form seedlings. This is the technique that is used to insert DNA constructs into chestnuts to create transgenic plants or to conduct gene edited plants (see later). Unfortunately, the efficiency of this process is low. We are engaged in discussions with groups using novel approaches to induce, transform, and regenerate embryogenic tissues.
As we become more proficient at transformation and regeneration, we also aim to add gene-editing technologies to our toolbox through collaboration with a group with proven success with plants beyond simple model systems. Our long-term strategy aims to increasingly use these techniques for an increasing number of experiments. Such an approach is not only faster and more targeted than conventional transgenic approaches, but also likely to be subject to less stringent regulation. Knocking out or enhancing expression of a candidate gene in pathogens or in chestnut represents a good early test of the value of the gene. The relative ease of the improved editing technologies will allow us to test many more genes or create many variants of single genes or promoters. Support for a project to develop skills and partner with us in gene-editing should be initiated in parallel or shortly after we obtain notable progress in efficient transformation and regeneration.
Resources
TACF has succeeded in developing reliable and steady income from more than 5,000 members and other generous donors. This, along with support from other organizations and competitive grants, has allowed TACF to support essentially all its current research activities, to continually update its major breeding facility at Meadowview Farm, and to employ a science team including three senior scientists with skills in quantitative genetics, genomics, bioinformatics, forest restoration, and project and facility management. The program is increasingly relying upon genomics and other new technologies that are benefiting from a number of new partnerships. TACF has been blessed with large numbers of enthusiastic member volunteers who serve as a unique and valuable resource as they support our horticultural and research efforts through our 16 state chapters. These volunteers are guided by two TACF-funded dedicated regional outreach coordinators and four regional research coordinators.
